Inputs to Dakota
Overview of Inputs
Dakota supports a number of command-line arguments, as described in Command-Line Options. Among these are specifications for the Dakota input file and, optionally, a restart file. Dakota input file syntax is described in the adjoining sections in Dakota Input File, with detailed keyword syntax in Keyword Reference. The restart file is described in Restarting Dakota.
A Dakota input file may be prepared with a text editor such as Emacs, Vi, or WordPad, or with the Dakota graphical user interface. The Dakota GUI is built on the Java-based Eclipse Framework [Ecl] and presents the Dakota input specification options in either a text editor view or a graphical view. It further includes templates and wizards for helping create Dakota studies and can invoke Dakota to run an analysis. Dakota GUI downloads for Linux, Windows, and Mac, are available from the Dakota website http://dakota.sandia.gov/, along with licensing information and installation tips. See Using Dakota GUI for more documentation.
Tabular Data Formats
The Dakota input file and/or command line may identify additional text files for tabular data import in contexts described in Data Imports. Examples include data from which to build a surrogate, points at which to run a list parameter study, post-run input data, and least squares and Bayesian calibration data. Dakota writes and reads tabular data with C++ stream operators/conversions, so most integer and floating point formats are acceptable for imported numeric data. Dakota supports the following tabular formats:
Annotated: In most contexts, Dakota tabular data defaults to “annotated” tabular format. An annotated tabular file is a whitespace-separated text file with one leading header row of comments/column labels. In most imports/exports, each subsequent row contains an evaluation ID and interface ID, followed by data for variables, or variables followed by responses, depending on context. This example shows 5 variables, followed by the 1 text_book response:
%eval_id interface TF1ln TF1ln hpu_r1 hpu_r2 ModelForm text_book 1 I1 0.97399 1.0476 12 4.133 3 14753 2 I1 0.94468 1.0636 4.133 12 3 14753 3 I1 1.0279 1.0035 12 4.133 3 14753
Another example is shown in Listing 26.
Note
Dakota 6.1 and newer include a column for the interface ID. See the discussion of custom-annotated format below for importing/exporting Dakota 6.0 format files.
For scalar experiment data files, each subsequent row contains an experiment ID, followed by data for configuration variables, observations, and/or observation errors, depending on context. This example shows 3 data points for each of two experiments.
%experiment d1 d2 d3 1 82 15.5 2.02 2 82.2 15.45 2
Free-form: When optionally specifying
freeformfor a given tabular import, the data file must be provided in a free-form format, omitting the leading header row and ID column(s). The raw num_rows x num_cols numeric data entries may appear separated with any whitespace including arbitrary spaces, tabs, and newlines. In this format, vectors may therefore appear as a single row or single column (or mixture; entries will populate the vector in order). This example shows the free-form version of the annotated data above:0.97399 1.0476 12 4.133 3 14753 0.94468 1.0636 4.133 12 3 14753 1.0279 1.0035 12 4.133 3 14753
Custom-annotated: In Dakota 6.2 and newer, a custom-annotated format is supported, to allow backward-compatibility with Dakota 6.0 tabular formats, which had a header and evaluation ID, but no interface ID. This can be specified, for example, with
method list_parameter_study import_points_file = 'dakota_pstudy.3.dat' custom_annotated header eval_idThe
custom_annotatedkeyword has options to controlheaderrow,eval_idcolumn, andinterface_idcolumn.
In tabular files, variables appear in input specification order as
documented in the reference manual. As of Dakota 6.1, tabular I/O has
columns for all of the variables (active and inactive), not only the
active variables as in previous versions. To import data corresponding
only to the active variables, use the keyword active_only when specifying the import file.
Note
Prior to October 2011, samples, calibration, and surrogate data
files were free-form format. They now default to annotated format,
though there are freeform and custom_annotated options. For
both formats, a warning will be generated if a specific number of
data are expected, but extra is found and an error generated when
there is insufficient data. Some TPLs like SCOLIB and JEGA manage
their own file I/O and only support the free-form option.
Data Imports
The Dakota input file and/or command line may identify additional files used to import data into Dakota.
AMPL algebraic mappings
As described in Algebraic Mappings, an AMPL specification of
algebraic input-to-output relationships may be imported into Dakota
and used to define or augment the mappings of a particular
interface. The files stub.nl, stub.row, and
stub.col define the mapping.
Genetic algorithm population import
Genetic algorithms (GAs) from the JEGA and SCOLIB packages support a
population import feature using the keywords initialization_type
flat_file = STRING. This is useful for warm starting GAs from
available data or previous runs. Refer to the flat_file keywords
in the method reference. The flat file must be in free-form
format.
Calibration data import
Calibration methods (deterministic least squares and Bayesian) require residuals, or differences between model predictions \(\mathbf{q}(\mathbf{\theta})\) and data \(\mathbf{d}\):
By default, if a Dakota input file specifies
responses-calibration_terms, the
simulation interface is required to return a vector of residuals
\(\mathbf{r}\) to Dakota. If in addition the input file includes
calibration_data or
calibration_data_file, Dakota
assumes the interface will return the model predictions
\(\mathbf{q}(\mathbf{\theta})\) themselves and Dakota will compute
residuals by differencing with the provided data.
There are two calibration data import mechanisms:
Scalar responses only with
calibration_data_file: This uses a single tabular text file to import data values and (optionally) experiment numbers, configurations, and observation variances. Each row of the data file expresses this information for a single experiment.Field and/or scalar responses with
calibration_data: In order to accommodate the richer structure of field-valued responses, this specification requires separate data files per response group (descriptor)DESC, per experimentNUM. The files are namedDESC.NUM.*and must each be in a tabular text format.
The tabular data files may be specified to be annotated (default),
custom_annotated, or freeform format.
Calibration data imports include the following information:
Configuration variables (optional): state variable values indicating the configuration at which this experiment was conducted; length must agree with the number of state variables active in the study.
Warning
In versions of Dakota prior to 6.14, string-valued configuration variables were specified in data files with 0-based indices into the admissible values. As of Dakota 6.14, strings must be specified by value. For example a string-valued configuration variable for an experimental condition might appear in the file as
low_pressurevs.high_pressure.Experimental observations (required): experimental data values to difference with model responses; length equal to the total response length (number of scalars + sum(field lengths)).
Experimental variances (optional): measurement errors (variances/covariances) associated with the experimental observations
For more on specifying calibration data imports, see the
nonlinear least squares examples and the reference
documentation for calibration_terms.
Note on variance: Field responses may optionally have scalar, diagonal, or matrix-valued error covariance information. As an example, Fig. 31 shows an observation vector with 5 responses; 2 scalar + 3 field (each field of length >1). The corresponding covariance matrix has scalar variances \(\sigma_1^2, \sigma_2^2\) for each of the scalars \(s1, s2\), diagonal covariance \(D_3\) for field \(f3\), scalar covariance \(\sigma_4^2\) for field \(f4\), and full matrix covariance \(C_5\) for field \(f5\). In total, Dakota supports block diagonal covariance \(\Sigma\) across the responses, with blocks \(\Sigma_i\), which could be fully dense within a given field response group. Covariance across the highest-level responses (off-diagonal blocks) is not supported, nor is covariance between experiments.
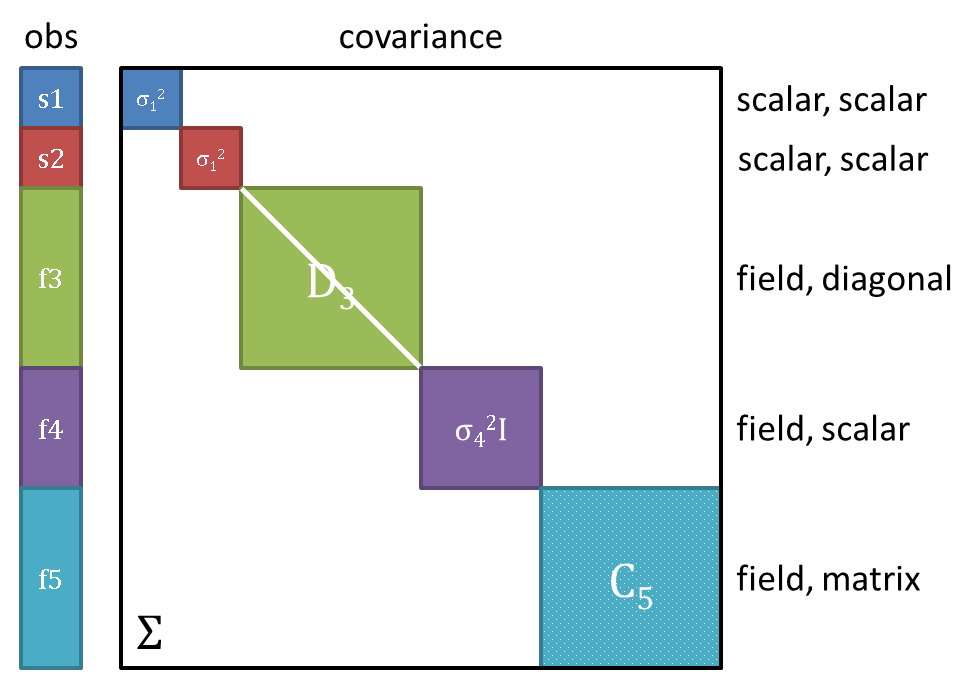
Fig. 31 An example of scalar and field response data, with associated block-diagonal observation error covariance.
PCE coefficient import
Polynomial chaos expansion (PCE) methods compute coefficients for
response expansions which employ a basis of multivariate orthogonal
polynomials. Normally, the polynomial_chaos method
calculates these coefficients based either on a spectral projection or
a linear regression (see Stochastic Expansion Methods). However, Dakota also
supports the option of importing a set of response PCE coefficients
from a file specified with import_expansion_file = STRING. Each
row of the free-form formatted file must be comprised of a coefficient
followed by its associated multi-index (the same format used for
output described in Stochastic expansion exports). This file import can be
used to evaluate moments analytically or compute probabilities
numerically from a known response expansion. Refer to
import_expansion_file for additional
information on this specification.
Surrogate Model Imports
Global data fit surrogates, including some stochastic expansions, may
be constructed from a variety of data sources. One of these sources is
an auxiliary data file, as specified by the keyword import_build_points_file. The file may be
in annotated (default), custom-annotated, or free-form format with
columns corresponding to variables and responses. For global
surrogates specified directly via keywords model surrogate global,
the keyword use_variable_labels will trigger validation and potential reordering of
imported variable columns based on labels provided in the tabular
header. Surfpack global surrogate models may also be evaluated at a
user-provided file containing challenge (test) points. Refer to the
global keywords for additional information on
these specifications.
Previously exported surfpack and experimental global surrogate models
can be re-imported when used directly in the global surrogate model
context. Importing from binary or text archive instead of building
from data can sometimes result in significant time savings with models
such as Gaussian processes. See the export_model and
import_model keywords in Keyword Reference for
important caveats on its use.
Variables/responses import to post-run
The post-run mode (supported only for sampling, parameter study, and DACE methods) requires specification of a file containing parameter and response data. Annotated is the default format (see :ref:input:tabularformat`), where leading columns for evaluation and interface IDs are followed by columns for variables (active and inactive by default), then those for responses, with an ignored header row of labels and then one row per evaluation. Typically this file would be generated by executing
dakota -i dakota.in -pre_run ::variables.dat
and then separate from daktoa adding columns of response data to
variables.dat to make varsresponses.dat. The file is
specified at the command line with:
dakota -i dakota.in -post_run varsresponses.dat::
To import post-run data in other formats, specify
post_run in the input file instead of on the
command-line, and provide a format option.

